Research
I am a trained population and statistical geneticist and have been a PhD student in Jeremy Berg’s lab in the department of human genetics at The University of Chicago since 2016. Before joining the Berg lab, I was an undergraduate researcher in the Graham Coop’s lab at UC Davis, supervised by Emily Josephs. Additionally, I completed summer undergraduate research in Eimear Kenny’s lab at the Icahn School of Medicine at Mount Sinai, supervised by Gillian Belbin.
My interests lie in understanding the complex interplay between genetics and the environment in shaping complex traits in humans. Thus far, my dissertation research has focused on using population genetic theory to model how confounding, either from environmental or background genetic effects, in GWAS generates bias in the distribution of polygenic scores that utilize those effect size estimates. Building off these theoretical results, I am developing methods to quantify how well existing approaches designed to control for confounding in GWAS perform under difference circumstances. In the future, I hope to continue working on problems surrounding polygenic prediction, specifically across diverse ancestry groups.
Human Statistical and Population Genetics
Confounding in polygenic scores
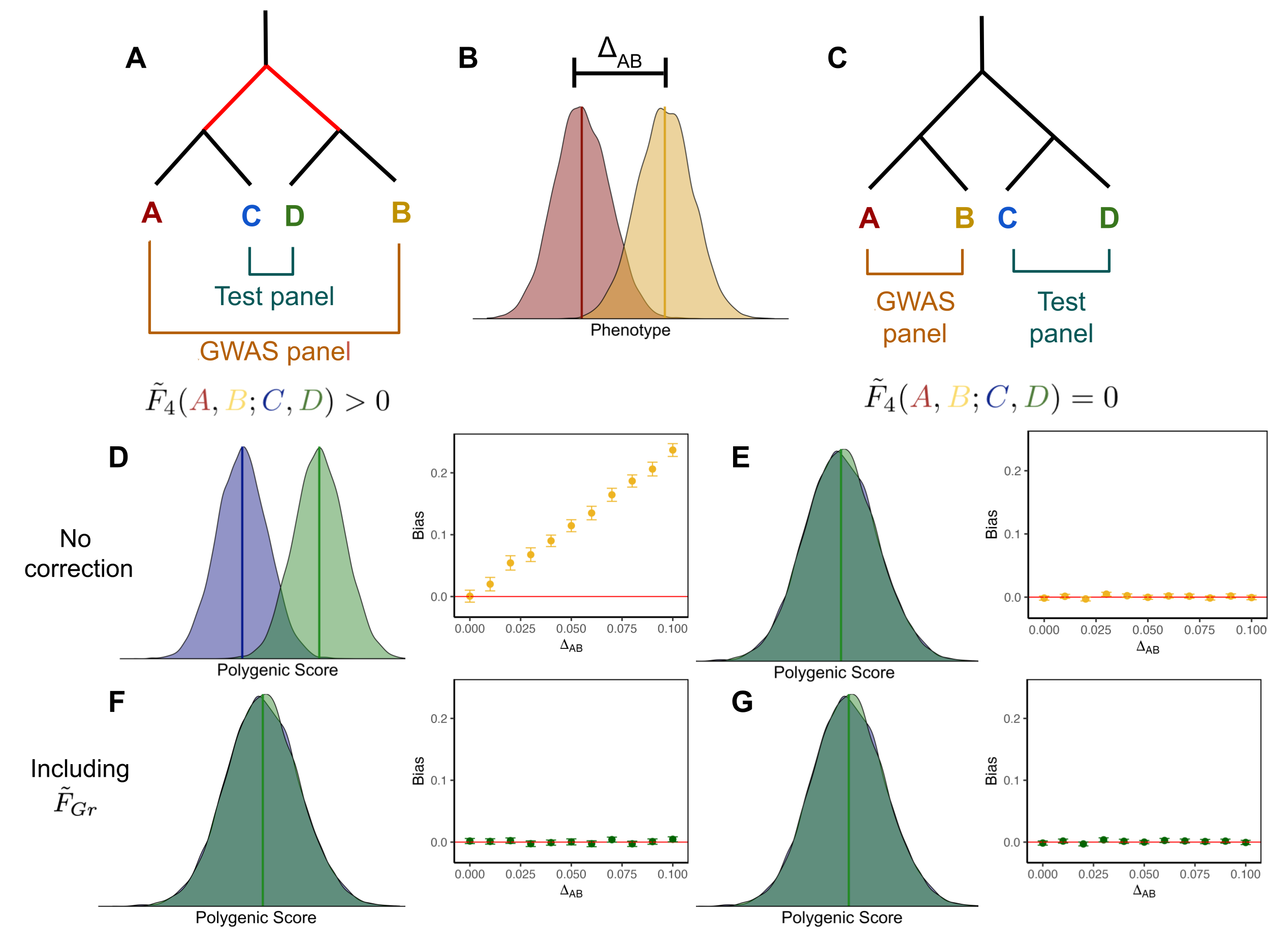
All of my dissertation work centers on the problem of confounding in polygenic scores. Complex traits are influenced by both genetics and the environment. Human geneticists increasingly use polygenic scores, calculated as the weighted sum of trait-associated alleles, to predict genetic effects on a phenotype. Differences in polygenic scores across groups would therefore seem to indicate differences in the genetic basis of the trait, which are of interest to researchers across disciplines. However, because polygenic scores are usually computed using effect sizes estimated using population samples, they are susceptible to confounding due to both the genetic background and the environment.
In our work we use theory from population and statistical genetics, together with simulations, to study how environmental and background genetic effects can confound tests for association between polygenic scores and axes of ancestry variation. We then develop a simple method to protect these tests from confounding, which we evaluate, alongside standard methods, across a range of possible situations. Our work helps clarify how bias in the distribution of polygenic scores is produced and provides insight to researchers wishing to protect their analyses from confounding.
Population structure in oceania
As a summer undergraduate intern in Dr. Eimear Kenny’s lab, I worked on a project inferring the demographic history of individuals living in Oceania using identity-by-descent. Specifically, I compared IBD sharing within and between islands. Generally, we found that inviduals living on islands farther from Euraisa had higher levels of IBD sharing and inviduals living on the coast shared more IBD with other islands than those living inland.
Evolution and Gene Expression in Maize
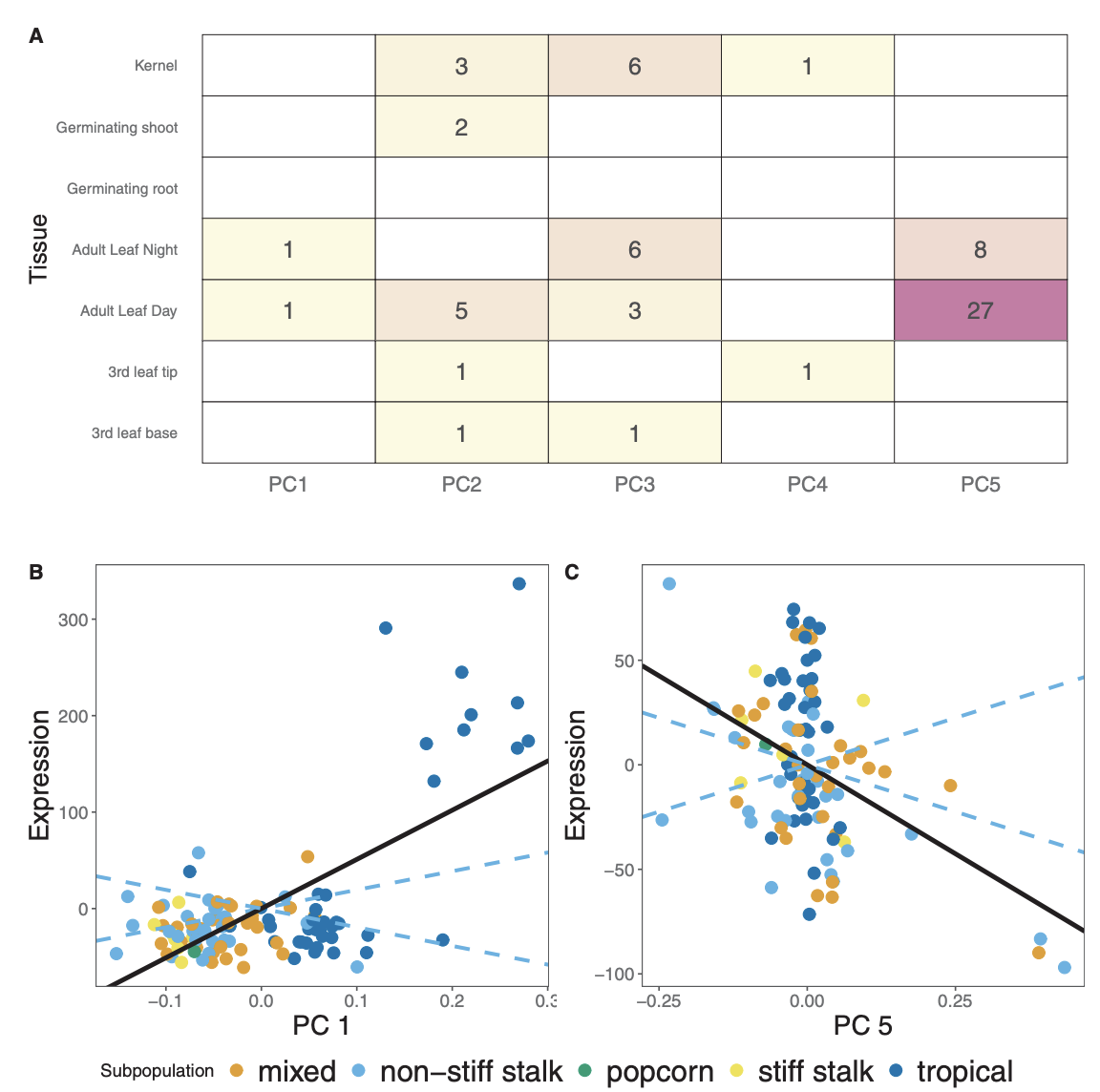
For my undergraduate honors thesis I worked with Emily Josephs in Graham Coop’s lab on a project studying the role of selection in shaping gene expression levels. We adapted a Qst - Fst framework to detect local adaptation for transcriptomes-wide gene expression levels in a population of diverse maize genotypes. We compared the number and types of selected genes across a wide range of maize populations and tissues, as well as selection on cold-response genes, drought-response genes, and coexpression clusters. We then identified a number of genes whose expression levels are consistent with local adaptation and showed that genes involved in stress response show enrichment for selection. Overall, this project sparked my interest in polygenic selection and the evolution of complex traits.